The laboratory conducts multiple research projects aimed at understanding microbial infections as complex systems at both systemic and molecular levels. These studies explore the interactions between pathogens, the host (biotic environment), and the surrounding abiotic environment to uncover novel pathogenicity mechanisms that can be targeted for therapeutic interventions. Additionally, the laboratory is actively involved in characterizing and developing new antimicrobial agents, with a particular focus on drug repurposing strategies.
1. Novel persistence determinants in chronic P. aeruginosa infections
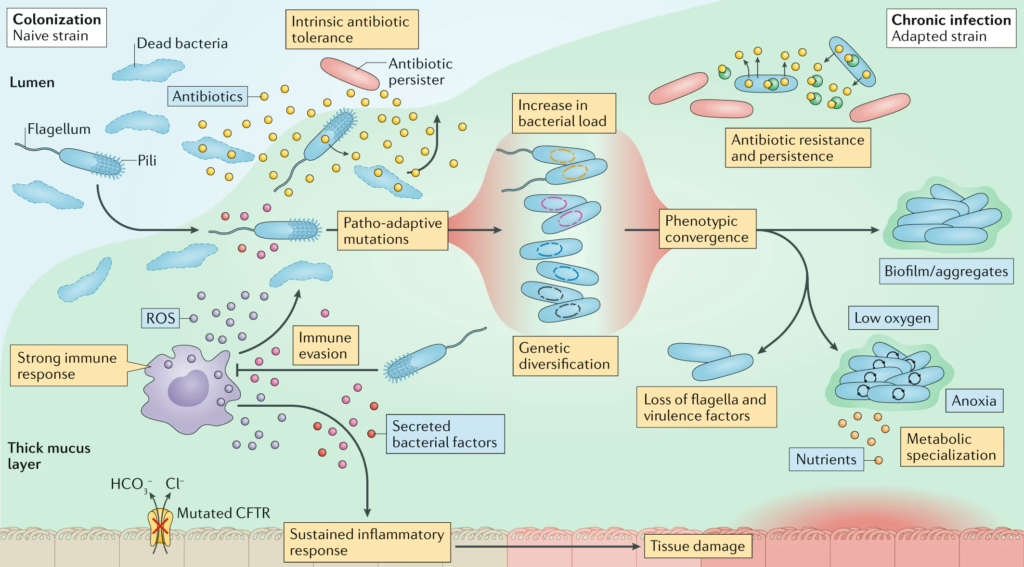
Cystic fibrosis (CF) is a genetic disorder characterized by the buildup of thick, sticky mucus in the lungs, which creates an environment conducive to persistent and often chronic bacterial infections. Among the pathogens most commonly associated with CF, Pseudomonas aeruginosa plays a dominant role due to its adaptability, ability to form biofilms, and resistance to conventional antibiotics. Infections caused by P. aeruginosa are a leading cause of morbidity and mortality in CF patients, making it a critical target for research and therapeutic development.
From https://doi.org/10.1038/s41579-020-00477-5
Our research leverages advanced metatranscriptomics to analyze samples derived from CF patients, enabling us to reconstruct the in vivo physiology of Pseudomonas aeruginosa during lung infections. This approach has revealed that a significant portion of the genes active during infection are poorly characterized or have entirely unknown functions, underscoring critical gaps in our understanding of this pathogen’s behavior in the host environment.
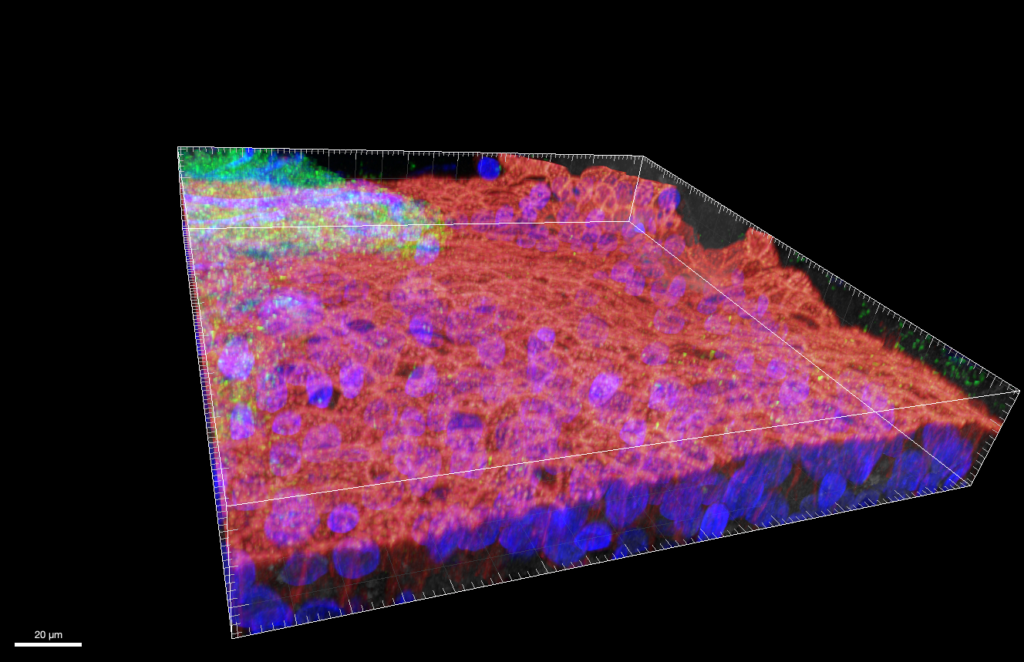
P. aeruginosa PA14 in fully-differentiated ALI coltures
Using a multidisciplinary approach that combines molecular biology, genomics, microbiology, cell biology and computational biology, we aim to unravel the roles of these enigmatic genes in the infection process. By linking gene activity to specific mechanisms of pathogenicity, we seek to identify novel determinants that drive P. aeruginosa virulence. Ultimately, our goal is to uncover innovative therapeutic targets that pave the way for new treatments, offering hope for improved outcomes in CF patients and reducing reliance on traditional antibiotics.
Related projects
Publications
2. Drug-repurpusing and antivirulence treatments as alternative to antibiotics
Our research focuses on developing innovative therapeutic strategies to address the growing threat of antibiotic resistance. Traditional antibiotics often target bacterial survival, leading to the emergence of resistant strains. In contrast, we aim to shift the paradigm by focusing on antivirulence therapies—a novel approach designed to disarm pathogens rather than kill them. By targeting key virulence factors essential for infection and host damage, antivirulence strategies reduce the selective pressure for resistance, offering a sustainable alternative to conventional treatments.
A central focus of our work is targeting bacterial cell metabolism, which plays a critical role in pathogenicity. We investigate how disrupting key metabolic pathways can impair a pathogen’s ability to thrive in the host environment. Specifically, we emphasize the use of nucleosides and nucleobase analogs, which have shown promising potential in interfering with bacterial pathogenesis.
Additionally, we explore drug repurposing to accelerate the discovery of effective antimicrobials. By identifying new therapeutic applications for existing drugs, we can bypass the lengthy and costly drug development process while uncovering compounds with unique mechanisms of action against microbial pathogens.
Our ultimate goal is to combine these approaches to design treatments that minimize resistance development, and provide effective solutions for managing persistent and multidrug-resistant infections.
Publications
- Ravishankar, S., Conte, A.L., Aliaga, S.J.C., Baldelli, V., Nielsen, K.L., Paroni, M., et al. (2025) The antimycotic 5-fluorocytosine is a virulence inhibitor of uropathogenic Escherichia coli and eradicates biofilm-embedded bacteria synergizing with β-lactams. Antimicrob agents Chemother e0028025.
- Ravishankar, S., Baldelli, V., Angeletti, C., Raffaelli, N., Landini, P., and Rossi, E. (2024) Fluoropyrimidines affect de novo pyrimidine synthesis impairing biofilm formation in Escherichia coli. Biofilm7: 100180.
- Rossi, E., Leccese, G., Baldelli, V., Bibi, A., Scalone, E., Camilloni, C., et al. (2022) Inactivation of the Pyrimidine Biosynthesis pyrD Gene Negatively Affects Biofilm Formation and Virulence Determinants in the Crohn’s Disease-Associated Adherent Invasive Escherichia coli LF82 Strain. Microorg10: 537.
- Rossi, E., Motta, S., Aliverti, A., Cossu, F., Gourlay, L., Mauri, P., and Landini, P. (2017) Cellulose production is coupled to sensing of the pyrimidine biosynthetic pathway via c‐di‐GMP production by the DgcQ protein of Escherichia coli. Environ Microbiol19: 4551–4563.
- Garavaglia, M., Rossi, E., and Landini, P. (2012) The Pyrimidine Nucleotide Biosynthetic Pathway Modulates Production of Biofilm Determinants in Escherichia coli. Plos One7: e31252.
- Antoniani, D., Rossi, E., Rinaldo, S., Bocci, P., Lolicato, M., Paiardini, A., et al. (2013) The immunosuppressive drug azathioprine inhibits biosynthesis of the bacterial signal molecule cyclic-di-GMP by interfering with intracellular nucleotide pool availability. Appl Microbiol Biot97: 7325–7336.
